PTGLweb structural motif overview
| Motif | Type | Motif abbreviation | Chains with motif in the database (of chains total) | Short info on motif |
|---|---|---|---|---|
| - | 4helix | 0 | 4helix info | |
| - | barrel | 0 | barrel info | |
| - | globin | 0 | globin info | |
| - | immuno | 0 | immuno info | |
| - | jelly | 0 | jelly info | |
| - | propeller | 0 | propeller info |
Motif info
Alpha motifs
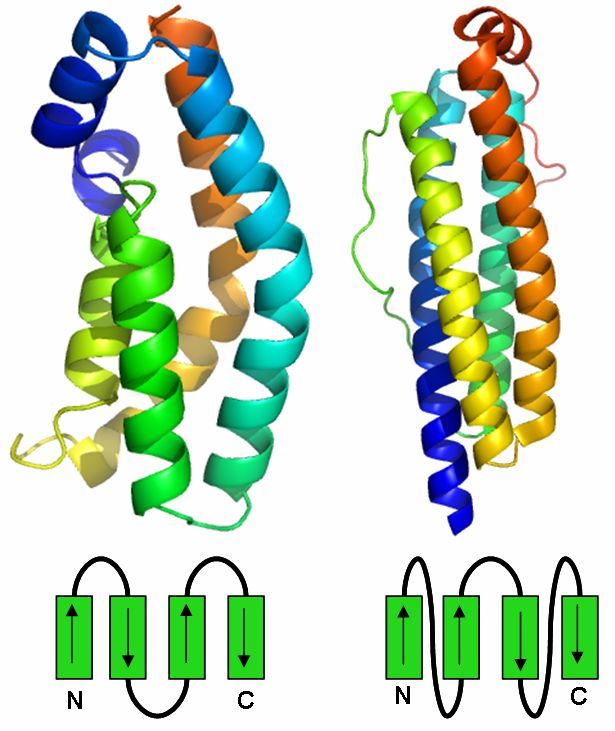
Four Helix Bundle
The Four Helix Bundle is a protein motif which consists of four alpha helices which arrange in a bundle.
There are two types of the Four Helix Bundle which differ in the connections between the alpha helices.
The first type of the Four Helix Bundle is all antiparallel and the second type has two pairs of parallel helices which have an antiparallel connection.
Found 0 times in the current database.
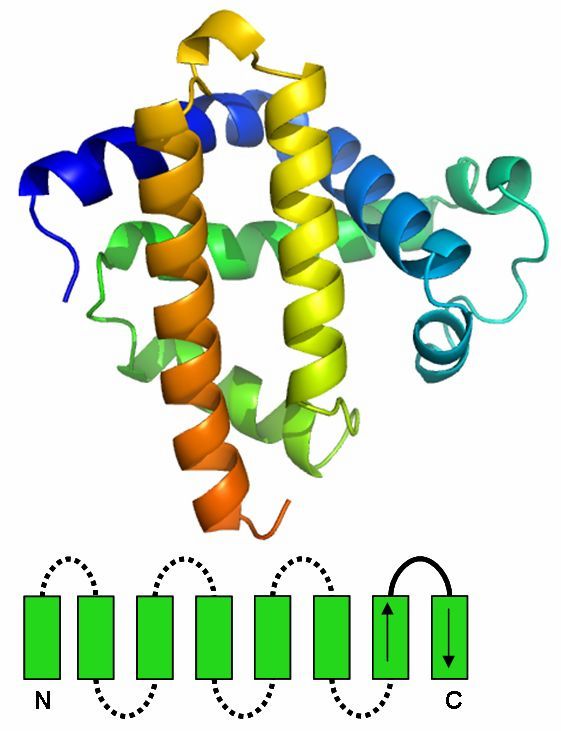
Globin Fold
The Globin Fold is an alpha helix structure motif which is composed of a bundle, consisting of eight alpha helices, which are connected over short loop regions.
The helices do not have a fixed arrangement, but the last two helices in sequential order are antiparallel.
Found 0 times in the current database.
Beta motifs
Up-and-down barrel
The up-and-down barrel is composed of a series of antiparallel beta strands which are connected via hydrogen bonds.
There are two major families of the up-and-down barrel, the ten-stranded and the eight-stranded version.
Found 0 times in the current database.
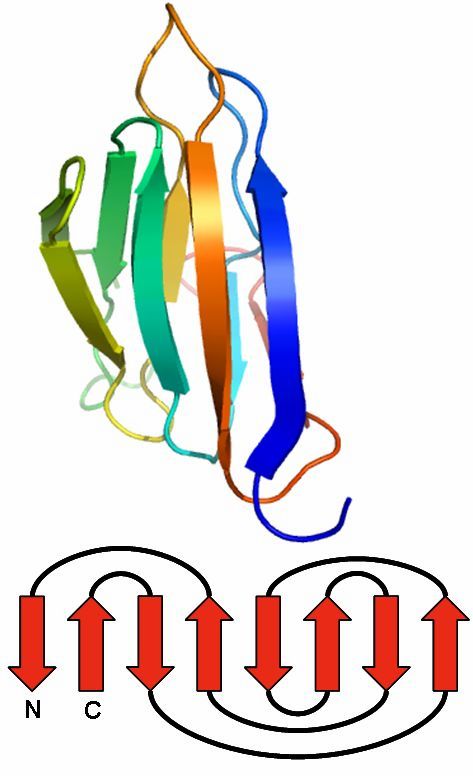
Immunoglobin fold
The immunoglobulin fold is a two-layer sandwich.
Usually, it consists of seven antiparallel beta strands, arranged in two beta sheets.
The first is composed of four and the second of three strands.
Both are connected via a disulfide bond to build the sandwich.
Found 0 times in the current database.
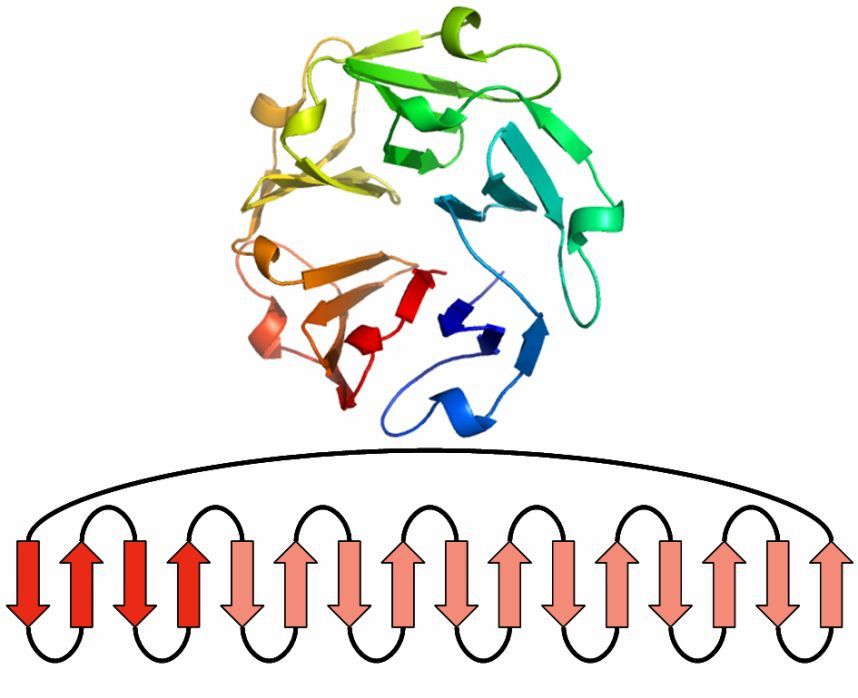
Beta Propeller
This beta motif contains between four and eight beta sheets, which are arranged around the center of the protein.
Each sheet is formed by four antiparallel beta strands.
One sheet makes up one of the propeller blades.
To build a four-bladed propeller, for example, four of these sheets are grouped together.
Found 0 times in the current database.
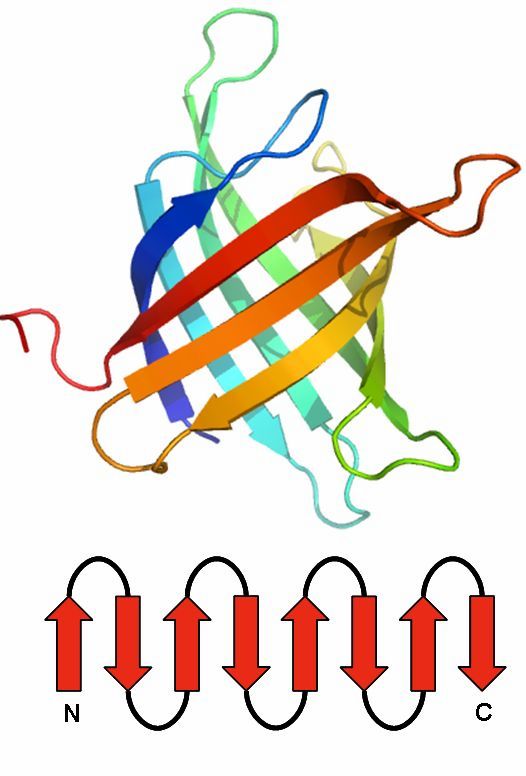
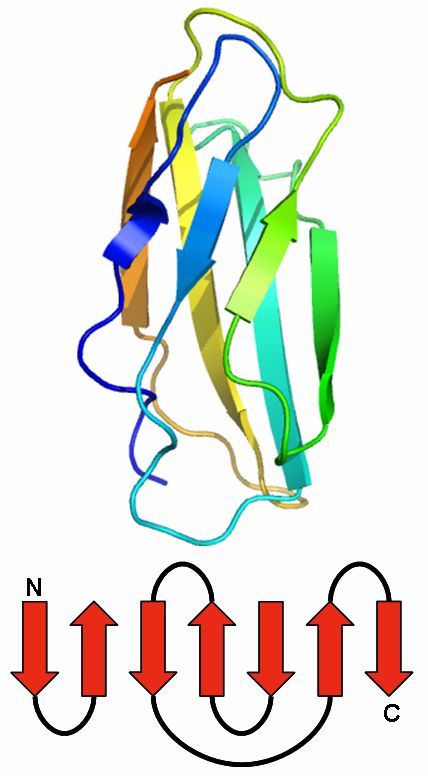
Jelly Roll
The Jelly Roll motif has a barrel structure, which seems like a jelly roll.
The barrel includes eight beta strands, which build a two-layer sandwich of four strands.
Found 0 times in the current database.