Step-by-step guide
Previous section
Biomedical examples
Table of contents
- Finding structurally similar chloroperoxidases
- Up-and-down beta-barrel
- Topology of central subunits of respiratory complex I
1. Finding structurally similar chloroperoxidases
Biomedical question
"Haloperoxidases catalyze the halogenation of organic compounds in the presence of halide ions and peroxides such as H2O2. They are named after the most electronegative halide they are able to oxidize." [Hofmann et al., 1998, J.Mol.B.]
- What is the structural topology of chloroperoxidase from Pseudomonas fluorescens?
- Which structures have a similar topology?
- With which protein classes are they associated?
Analysis with PTGL
The chloroperoxidase from Pseudomonas fluorescens can be found in the Protein Data Bank via the accession code 1a8s [Hofmann et al., 1998, J.Mol.B].
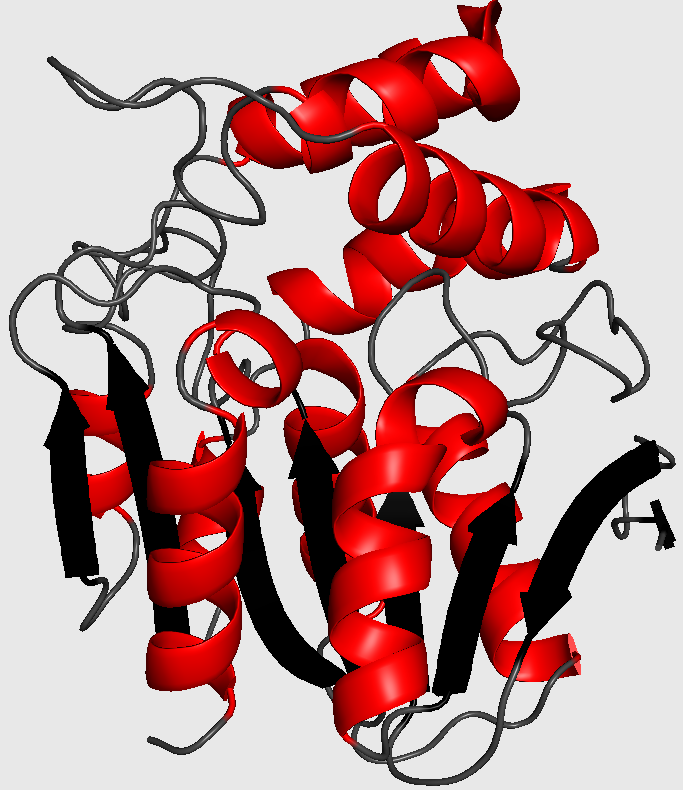
Cartoon-style three-dimensional structure of chloroperoxidase from P. fluorescens (1a8s). The chains are colored grey, black and red for coils, strands and helices, respectively. Ligands are drawn as sticks and colored magenta.
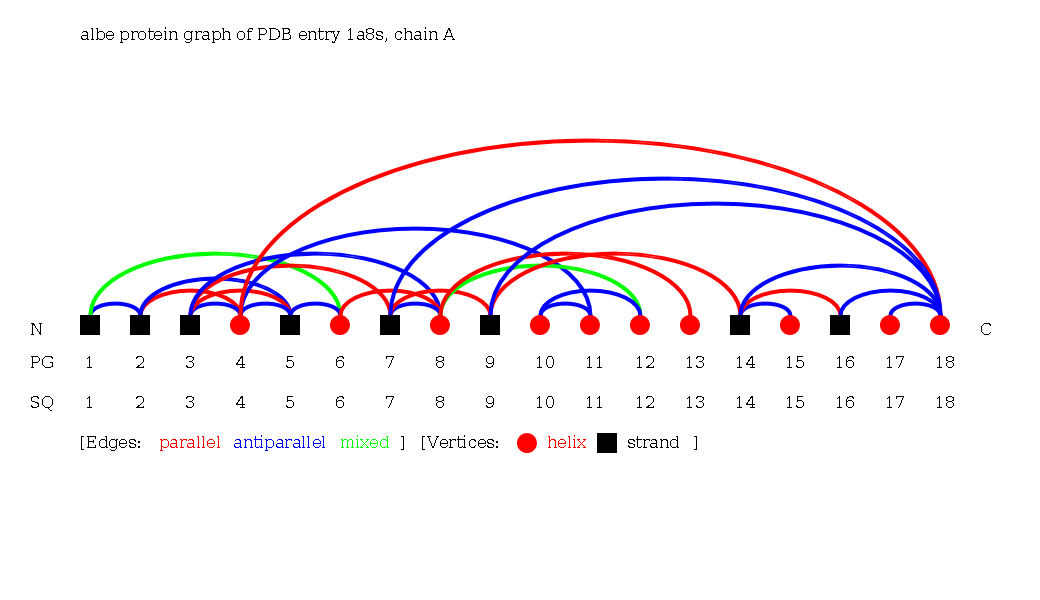
- Search for the Protein Graph of chloroperoxidase from Pseudomonas fluorescens (1a8s):
Switch to the beta topology:
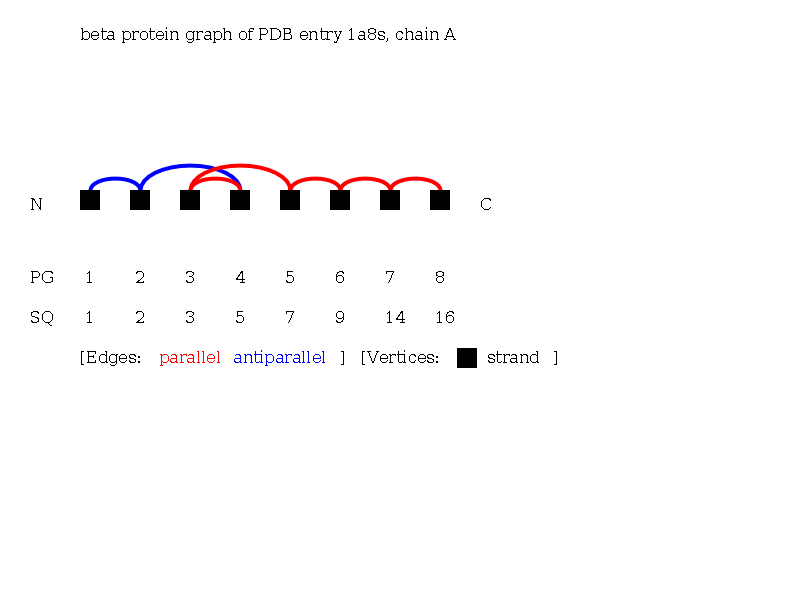
Go to the Folding Graph and retrieve the reduced linear notation of the beta Folding Graph of 1a8sA:
- Use the linear notation search for the beta reduced linear notation.
- Browse the results and look for the field 'classification' for each protein.
Answer
- The protein exhibits an alpha beta class topology with strands and helices. The vertices of the strands are connected across one path of edges, and therefore the strands form a single sheet. The helices are not in spatial neighborhood and show no characteristic topology. The beta Protein Graph shows the topology of the alpha-beta hydrolase fold [Homquist, 2000, Curr. Protein Pept. Sc.].
- The linear notation search yields 2,078 protein chains with this topology, e.g., chloroperoxidase T from Kitasatospora aureofaciens (1a7u), chloroperoxidase L from Streptomyces lividans (1a88), bromoperoxidase A1 from Kitasatospora aureofaciens (1a8q).
- The first 100 results show the following functional classifications that all exhibit the same beta topology: haloperoxidase, aminopeptidase, hydrolase, dehalogenase, immune system, hydrolase/hydrolase inhibitor, transferase, and serine hydrolase.
2. Up-and-down beta-barrel
Biomedical question
„The up-and-down beta-barrel is a common folding motif found frequently in proteins that bind and transport hydrophobic ligands. [...] Proteins belonging to this class of up-and-down beta-barrels are found typically to be lipid-binding proteins in which the interior surface forms a cavity or pit that serves as the ligand binding region.“ [LaLonde et al., 1994, FASEB J.]
- Which PDB structures contain an up-and-down beta-barrel fold?
- Which of them contain a ligand in the interior of the beta-barrel?
Analysis with PTGL
- Use the motif search for the up-and-down beta-barrel.
- Open beta-ligand Protein Graphs of found chains, e.g., chain A from retinol-binding protein from Sus scrofa domesticus (1aqb).
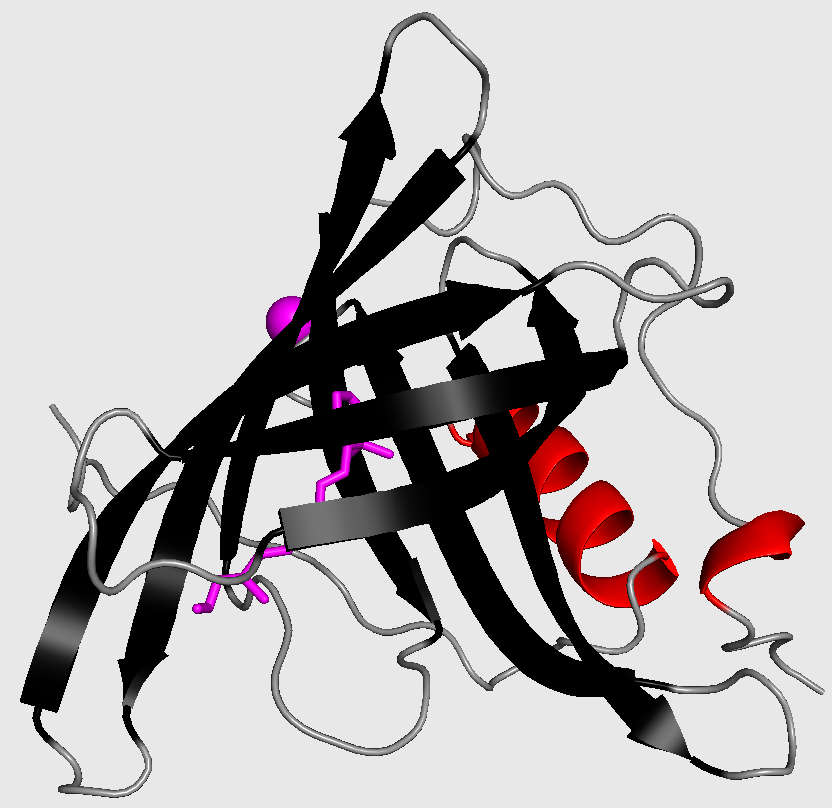
Cartoon-style three-dimensional structure of retinol-binding protein from Sus scrofa domesticus. Ligands are drawn as sphere or sticks.
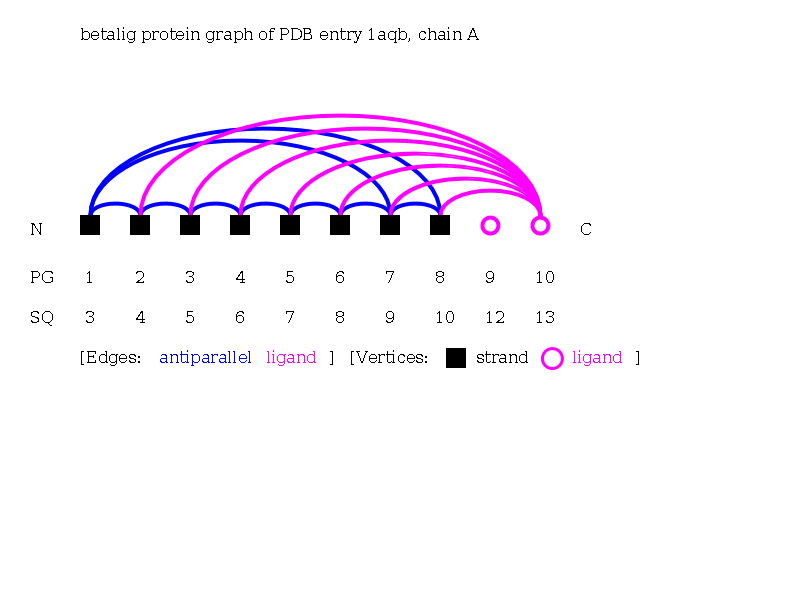
Answer
- The motif search yields 0 protein chains, e.g., odorant binding protein from nasal mucosa from Sus scrofa (1a3y), retinol-binding protein from Sus scrofa domesticus (1aqb), egg-white avidin from Gallus gallus (1avd), egg-white apo-avidin from Gallus gallus 1ave, beta-lactoglobulin from Bos taurus (1b8e).
- The up-and-down beta-barrels motif can be seen in the Protein Graphs by numerous strands exhibiting a cycle of antiparallel edges. A ligand lies within the center of the barrel if it is connected with multiple up to all strands of a barrel, e.g., 1aqbA, 1brpA, 1bsoA, 1bxwA.
3. Topology of central subunits of respiratory complex I
Biomedical question
„With a molecular mass of about 1 MDa, mitochondrial complex I is the largest multi-subunit complex of the respiratory chain. Complex I couples electron transfer from NADH to ubiquinone with transmembrane proton pumping contributing to the proton motive force used for ATP synthesis. [...] Complex I contributes significantly to the formation of mitochondrial reactive oxygen species (ROS) and is implicated in the pathogenesis of numerous hereditary and degenerative disorders [...] Complex I comprises 14 central subunits that execute the core bioenergetic functions and that are conserved from bacteria to humans. [...] The mass of the mitochondrial enzyme is almost double compared to its bacterial counterpart, because it comprises some 30 accessory subunits.“ [Wirth et al., 2016, BBA. Bioenergetics]
- What is the topology of the conserved core of human's respiratory complex I?
- Is the topology of the central subunits conserved across complex I of Thermus thermophilus?
Analysis with PTGL

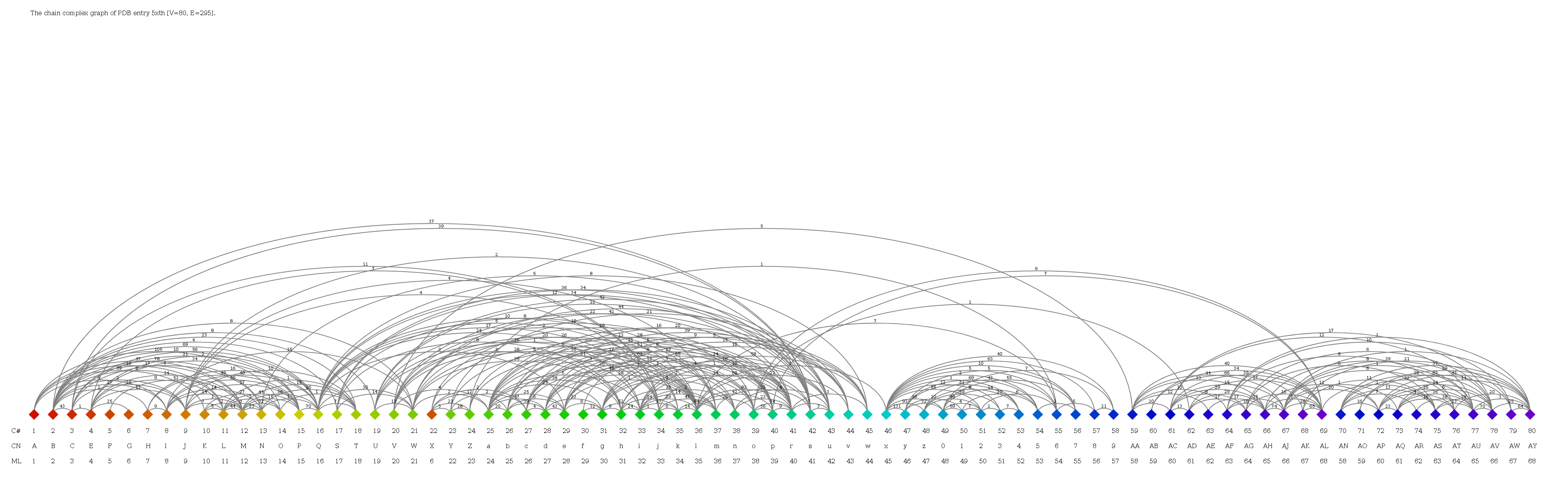
C-alpha trace of three-dimensional structure of respiratory supercomplex I1III2IV1 from H. sapiens (5xth). Chains are colored individually.
- Use the Complex Graph search for human respiratory supercomplex I1III2IV1 (5xth).
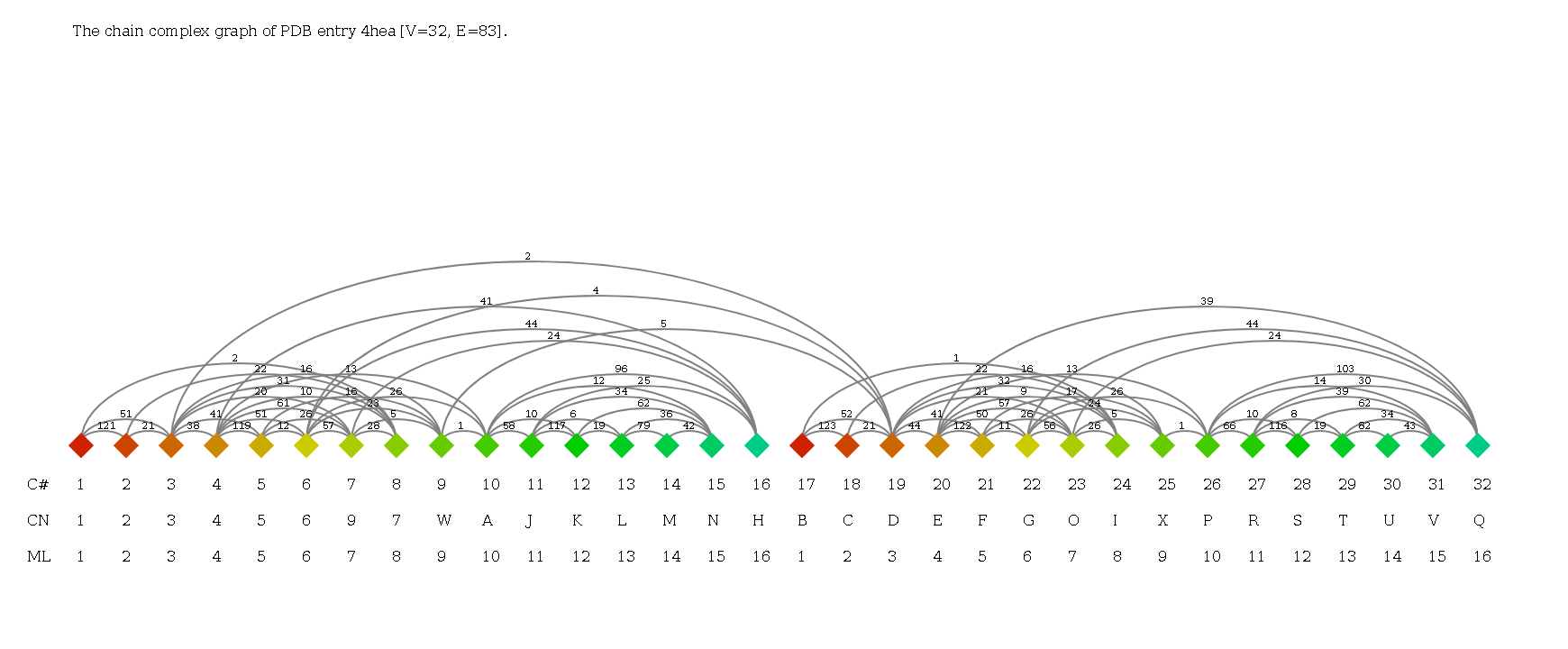
- Use the Complex Graph search for complex I of T. thermophilus (4hea).
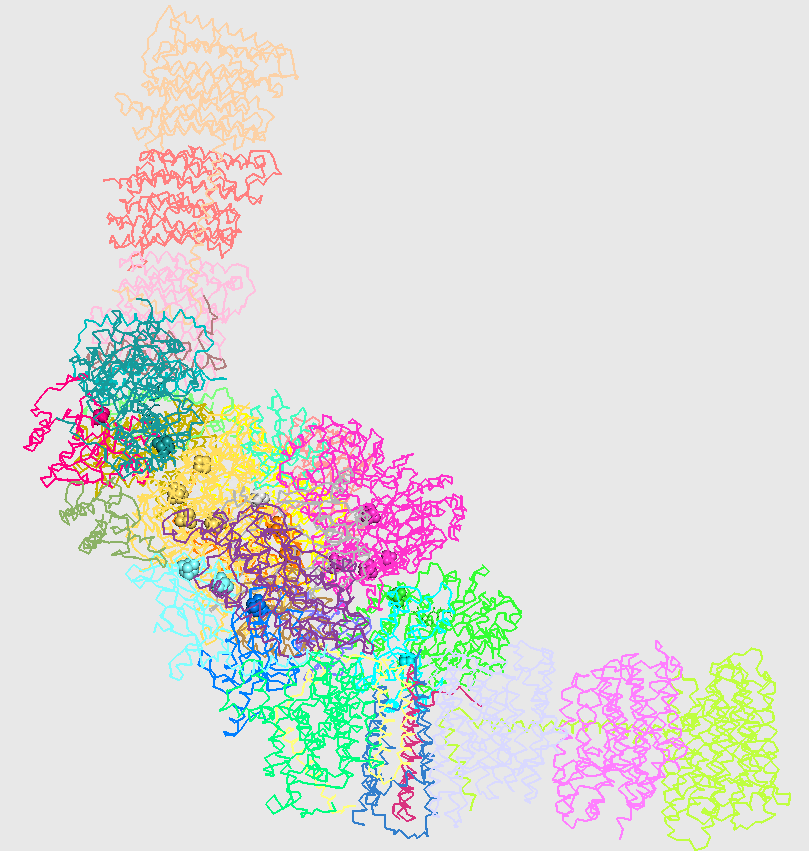
C-alpha trace of three-dimensional structure of two copies of respiratory complex I from T. thermophilus (4hea). Chains are colored individually.
Central subunits of respiratory complex I and their chain names for H. sapiens and T. thermophilus. Subunits of the same row are homologous. [Wirth et al., 2016, BBA. Bioenergetics]
| Homo sapiens (5xth) | Thermus thermophilus (4hea) | ||
|---|---|---|---|
| Subunit | Chain ID | Subunit | Chain ID |
| NDUFS1 | M | Nqo3 | 3 |
| NDUFV1 | A | Nqo1 | 1 |
| NDUFS2 | J | Nqo4 | 4 |
| NDUFS3 | P | Nqo5 | 5 |
| NDUFV2 | O |
Nqo2 | 2 |
| NDUFS8 | B | Nqo9 | 9 |
| NDUFS7 | C | Nqo6 | 6 |
| NU1M (ND1) | s | Nqo8 | H |
| NU2M (ND2) | i | Nqo14 | N |
| NU3M (ND3) | j | Nqo7 | A |
| NU4M (ND4) | r | Nqo13 | M |
| NU5M (ND5) | l | Nqo12 | L |
| NU6M (ND6) | m | Nqo10 | J |
| NULM (ND4L) | k | Nqo11 | K |
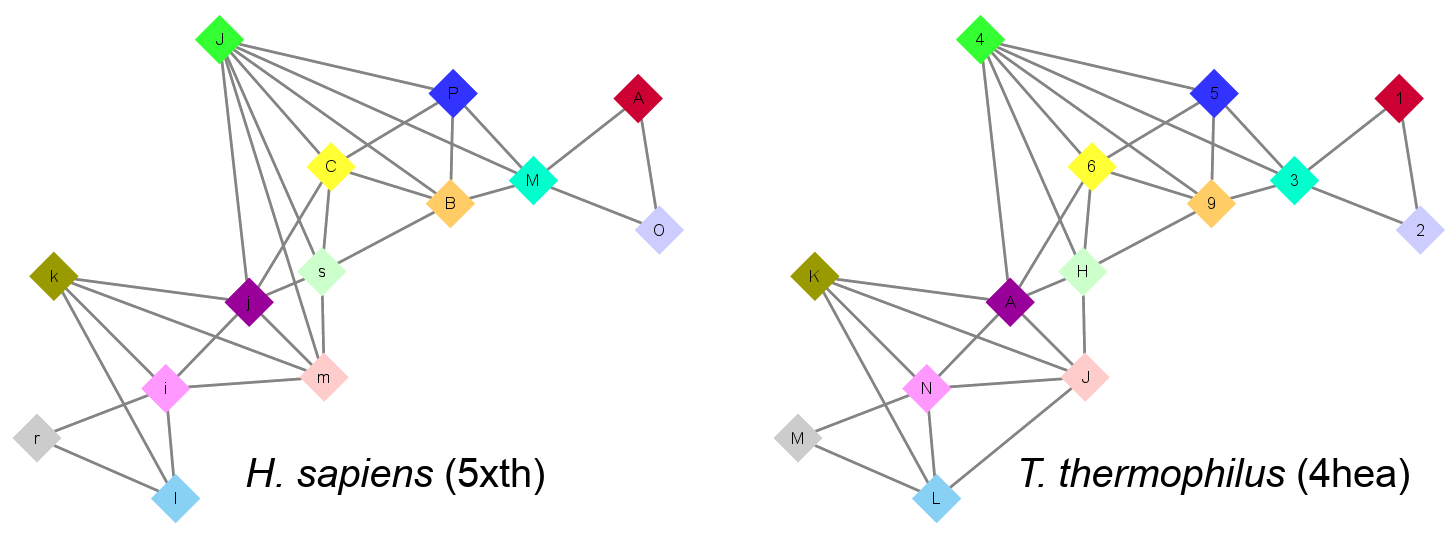
Answer
- NDUFS2 (J) is a key subunit in contact with seven other subunits: NDUFV1 (A), NU6M (m), NU1M (s), NDUFS7 (C), NDUFS8 (B), NDUFS1 (M), NDUFS3 (P). NDUFV1 (A) and NDUFV2 (O) are peripheral, only connected with each other and with NDUFS1 (M). NU4M (r) is also peripheral as it is only in contact with NU2M (i) and NU5M (l). Other subunits are more central with numbers of contacts between three and five.
- The topology of the central subunits is fully conserved with one exception. In H. sapiens, there is a contact between NU6M (m) and NDUFS2 (J) while in T. thermophilus, there is a contact between the homologue of NU6M, Nqo10 (J), and Nqo12 (L).
Next section
Help