PTGLweb is a web server and database of protein structure topologies [Wolf et al., 2020, Bioinformatics; Koch and Schäfer, 2018, Current Opinion in Structural Biology; Schäfer et al., 2015, Bioinformatics]. PTGLweb is based on topology graphs of different levels of abstraction computed by PTGLtools (formerly labeled VPLG). The graphs can be automated computed, visualized and used for further analysis.
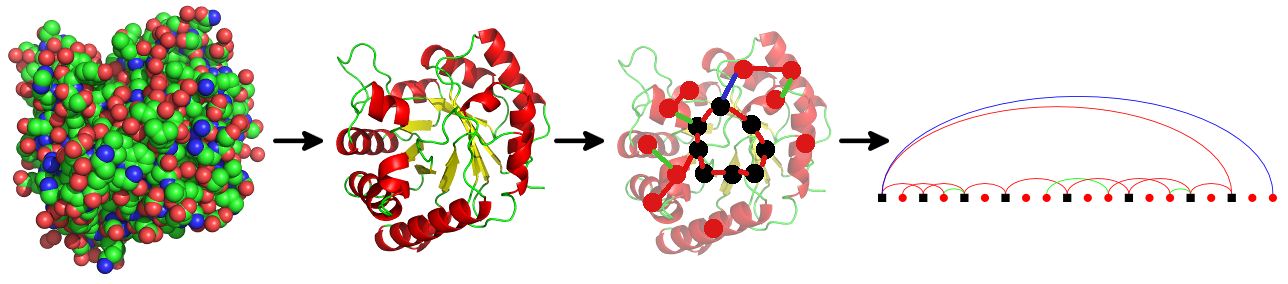
This web server allows you to search for protein motifs which can be detected in the graphs. It also provides standardized 2D visualizations of protein graphs and folding graphs. In contrast to the manually curated CATH and SCOP databases, the method used by this server is fully automated. Similar servers, which also support substructure search, include ProSMoS and Pro-Origami.